Open Access |Peer-reviewed |Review Article
Syed Hussain Basha*
Innovative Informatica Technologies, Hyderabad - 500049, India.
Published: April 01, 2020 DOI: 10.5281/zenodo.3747641
Abstract
A novel Coronavirus (CoV), SARS-CoV-2 surfaced in late 2019 in Wuhan, China. SARS-CoV-2, unlike previously identified coronaviruses responsible for severe acute respiratory syndrome (SARS) outbreak in 2002 and Middle East Respiratory Syndrome (MERS) in 2012, has rapidly spread across the globe claiming thousands of lives. The new disease caused by SARS-CoV-2 is named coronavirus disease 2019 (COVID-19) by World Health Organization (WHO). As the SARS-CoV-2 is making its way around the world, researchers and doctors are in search of drugs to treat and stop the spread of the disease. Several drugs such as hydroxychloroquine / azithromycin, chloroquine, remdesivir, Ritonavir / lopinavir and favipiravir are currently undergoing clinical trials to test their ability to treat COVID-19 without adverse effects to the infected patients. However, there are many other small compounds being reported from across globe with potential to treat COVID-19 infected patients worth considering for further studies. In this scenario, this review aims to summarize the past, present and future of compounds that can be used to treat corona virus, highlighting compounds presently in clinical trials and all the way to final stages of bringing a drug to market.
Keywords: SARS-CoV-2, COVID-19, anti-coronavirus drugs, 2019‐CoV, Coronaviridea.
| Citation: Syed Hussain Basha (2020) Corona virus drugs – a brief overview of past, present and future, Journal of PeerScientist 2(2): e1000013. |
| Received: March 21, 2020; Accepted April 01, 2020; Published April 01, 2020. |
| Copyright:© 2020 Syed Hussain Basha. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. |
| Data Availability: All relevant data are within the paper and its Supporting Information files. |
| Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. |
| Competing interests: The authors have declared that no competing interests exist. |
| *E-mail: shb@innovativeinformatica.com; hassainbasha53@gmail.com | Phone: +91-9177247605 |
Introduction
In late 2019, sudden rise in respiratory related disease cases in china triggered the identification of its source as a novel corona virus, termed Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2). The disease caused by novel SARS-CoV-2 is named as COVID-19 by World Health Organization (WHO) [1]. The culprit virus belongs to the Coronaviridea family of corona viruses that caused two other outbreaks, namely Severe Acute Respiratory Syndrome (SARS) in 2002 [2] and Middle East Respiratory Syndrome (MERS) in 2012 [3]. After its initial outbreak in Wuhan, China; it has rapidly spread across globe infecting population of 192 countries (figure 1). As of March 29th 2020, according to WHO official records there are 697,609 infected people globally, far surpassing 10,600 combined total cases of SARS and MERS. Out of these 614,136 infected people 181,563 cases were closed with 148,447 recovered and 33,116 deaths (figure 2), with USA leading with 142,047 cases, followed by Italy 97,689 and China with 81,439. Details about laboratory-confirmed number of infected and dead people in top 10 COVID-19 infected countries are shown in figure 3.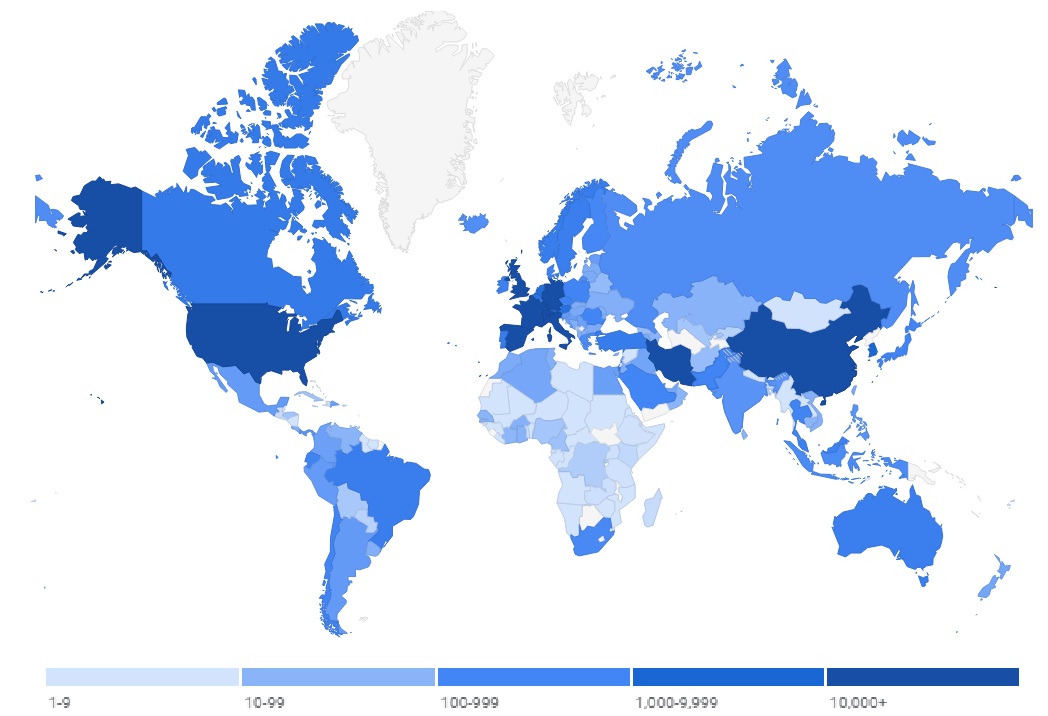 Figure 1: World map showing the infected countries of probable exposure of laboratory-confirmed SARS-CoV-2 cases. Color intensity signifies the number of infected people in the respective country. Map credits: Google crisis response dashboard (https://google.org/crisisresponse/covid19-map) based on World Health Organization database.
Figure 1: World map showing the infected countries of probable exposure of laboratory-confirmed SARS-CoV-2 cases. Color intensity signifies the number of infected people in the respective country. Map credits: Google crisis response dashboard (https://google.org/crisisresponse/covid19-map) based on World Health Organization database. Figure 2: Info graph depicting COVID-19 key statistics such as total number of lab-confirmed SARS-CoV-2 infected patients, recovered and total deaths registered between January 22nd and March 28th 2020 as per WHO official records.
Figure 2: Info graph depicting COVID-19 key statistics such as total number of lab-confirmed SARS-CoV-2 infected patients, recovered and total deaths registered between January 22nd and March 28th 2020 as per WHO official records.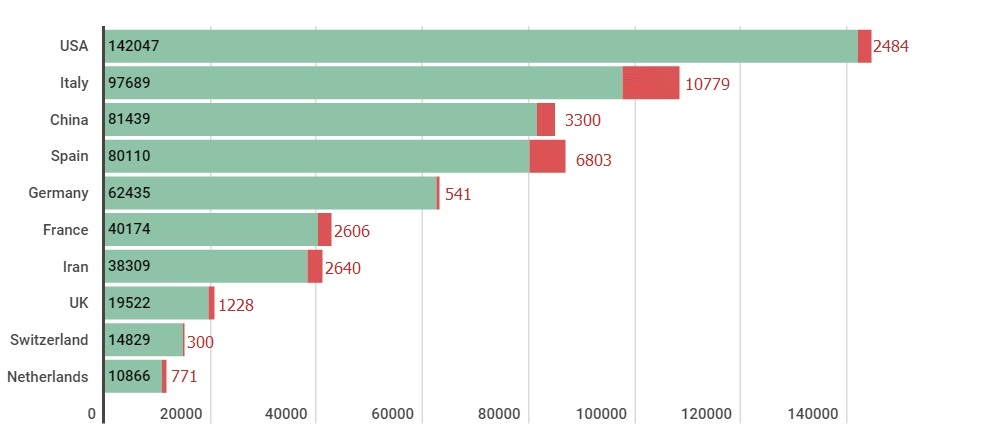 Figure 3: Details about number of laboratory-confirmed infected and deaths of top 10 COVID-19 infected countries as per WHO official records. X-axis shows the total number of registered cases. Y-axis shows the bar charts of each country.
Figure 3: Details about number of laboratory-confirmed infected and deaths of top 10 COVID-19 infected countries as per WHO official records. X-axis shows the total number of registered cases. Y-axis shows the bar charts of each country.
Overall, there are atleast 39 corona viruses researchers are familiar with [4], among which seven are most commonly found infecting humans:
- 229E (alpha coronavirus)
- NL63 (alpha coronavirus)
- OC43 (beta coronavirus)
- HKU1 (beta coronavirus)
- MERS-CoV (the beta coronavirus that causes Middle East Respiratory Syndrome, or MERS)
- SARS-CoV (the beta coronavirus that causes severe acute respiratory syndrome, or SARS)
- SARS-CoV-2 (the novel coronavirus that causes coronavirus disease 2019, or COVID-19) [5].
SARS-CoV-2 is a single stranded, positive strain RNA virus with a protein shell and membrane [6]. Due to their dependency on hosts’ molecular machinery to replicate themselves, these RNA based viruses are prone to mutations when they transfer from one animal to another. When infection transfers from animal to human, they can evolve into new virus like SARS-CoV, MERS-CoV [7] and SARS-CoV-2. Genetic information of the virus is the starting point in understanding the origin of the pathogen and one of the valuable information in designing strategies to fight against it.
As soon as the genetic makeup of SARS-CoV-2 is revealed by Li-Li Ren et.al on January 25th 2020 [8], researchers across globe began comparing the novel coronavirus’s genome with SARS and MERS to determine if any of the drugs developed against SARS and MERS can work against this novel SARS-CoV-2. Due to this comparative study, few potential drug targets of SARS-CoV-2 have been identified. Among several, human angiotensin-converting enzyme 2 (hACE2) [9], papain-like protease (PL pro) [10], 3C-like protease (3CL pro) [11], RNA-dependent RNA polymerase (RdRp) [12], helicase [13] and N7 methyltransferase [14] are some of primary drug targets. While, Human dipeptidyl peptidase IV (DDP4) [15], receptor-binding domain (RBD) [16], protease cathepsin L [17], type II transmembrane serine protease or TMPRSS2 [18], membrane-associated replication and transcription complex (RTC) [19], ADP-ribose-100-phosphatase [20], primase [21], exonuclease [22], endoribonuclease [23] and endoplasmic reticulum–Golgi intermediate compartment (ERGIC) [24] are some of other important drug targets against coronavirus. According to an initial analysis among SARS-CoV-2 infected people, about 80.9% show only mild flu like symptoms such as fever and cough and can be recovered at home. However, with good prognosis, 13.8% infected can develop these mild symptoms into severe cases of pneumonia and acute respiratory failure. About 4.7% are at risk of septic shock, respiratory or multi-organ failure with about 2% fatality rate and relatively very few cases are reported among children [25]. As the SARS-CoV-2 making its way around the world, researchers and doctors are in search for drugs to treat and stop the spread of the disease. Since, there are no specific therapeutic options available at present, health officials are primarily relying on quarantining the infected to contain the virus spread and repurposing already existing anti-viral drugs and antibiotics to treat infection based on symptoms. Thus, there remains an urgent need for the discovery / development of SARS-CoV-2 specific antiviral therapeutics and vaccines. In this scenario, this review aims to summarize the past, present and future of drugs that can be used to treat corona virus. Highlighting compounds presently in clinical trials and all the way to final stages of brining a drug to market. Although there are many potential peptides, recombinant proteins, monoclonal antibodies and many vaccines under development, this review is limited to the discussion of small compounds only.
Past:
Chloroquine is a 9-aminoquinoline, a well known compound since 1934 with interesting biochemical properties and has been explored against several viral infections. In 2003 Andrea Savarino et.al, suggested that it is worth evaluating Chloroquine / hydroxychloroquine against 2002 SARS outbreak characterized by symptoms associated with immune-hyperactivation and / or inflammatory processes [26]. In 2005 Martin J Vincent et.al, has took the study further and reported that chloroquine has strong antiviral effects on SARS-CoV infection. Associated mode of action is via interfering with terminal glycosylation of the cellular receptor, angiotensin in converting enzyme 2, which was thought to negatively influence the virus-receptor binding and abrogated the infection [27].
In 2003, J Cinatl et.al, identified glycyrrhizin, an active component of liquorice roots as active against inhibiting SARS-associated virus replication after assessing 6-azauridine, mycophenolic acid, ribavirin, pyrazofurin and glycyrrhizin against two coronavirus from patients diagnosed with SARS at clinical centre of Frankfurt University, Germany [28]. Yuen et al. in 2004 disclosed anti-SARS-CoV activity of Baicalin – A Chinese medicinal herbal compound with EC50 of 12.5 – 25 μg/ml at 48 h and 25 – 50 µg/ml at 72 h, CC50 of > 100 μg/ml and SI of > 8 with unknown mechanism of action [29]. In 2003, Kanchan Anand et.al, determined crystal structure main proteinase (Mpro) of human coronavirus (strain 229E). From the structural analysis, they concluded that AG7088 compound derivatives as a good starting point for the design of anti-coronaviral drugs [30]. Wu CY et.al, in 2004 identified that one of their compound named Compound (2) which was actually developed as a transition-state HIV-1 protease inhibitor turned out to be a potential SARS-CoV 3CLpro inhibitor with EC50 of 3 μM [31]. In 2005, Jason Paragas et.al, reported potent antiviral activity of interferon alfacon1 against SARS-CoV with an IC50 of 0.001 µg/ml as evaluated in a cytopathic effect protection (CPE) assay [32]. Following virtual screening of 8,000 existing drugs by a docking approach against a homology model of SARS-CoV 3CLpro, Cinanserin (SQ 10,463) – a serotonin antagonist was chosen for further experimental evaluation. Furthermore, binding of cinanserin with bacterially expressed 3CLpro of SARS-CoV and the related human coronavirus 229E (HCoV-229E) was demonstrated by surface plasmon resonance technology, with enzymatic catalytic activity 50% inhibitory concentration (IC50) values estimated as 5 µM [33]. In 2008, Qingang Yang et.al, designed and synthesized cinanserin analogs and tested for the inhibitory activities against SARS-coronavirus (CoV) 3CLpro by fluorescence resonance energy transfer (FRET) assay. Four analogs compounds 10, 13, 26 and 27 show significant activities, especially compound 26 with an IC50 of 1.06 mM [34].
In 2009, Xin chen et.al, reported Thiopurine analogue inhibitors - 6-thioguanine (6TG) and 6-mercaptopurine (6MP) as specific inhibitors for the papain-like protease (PLpro) of SARS—coronavirus (CoV) [35]. In 2009, L. A. Baltina et.al, summarized antiviral activity of glycyrrhizic acid (GA) and its derivatives and emphasized on chemical modification of GA as a promising way of designing new highly active antiviral drugs [36]. Several other compounds which might have potential anti-coronavirus activity have been summarized by Tong TR in 2009 [37]. In 2010 Falgun Shah et.al, summarized computational approaches used for the design of SARS-CoV Mpro. A wide range of structure and ligand based modeling strategies have been discussed with focus on important features of ligand to be recognized by drug target [38].
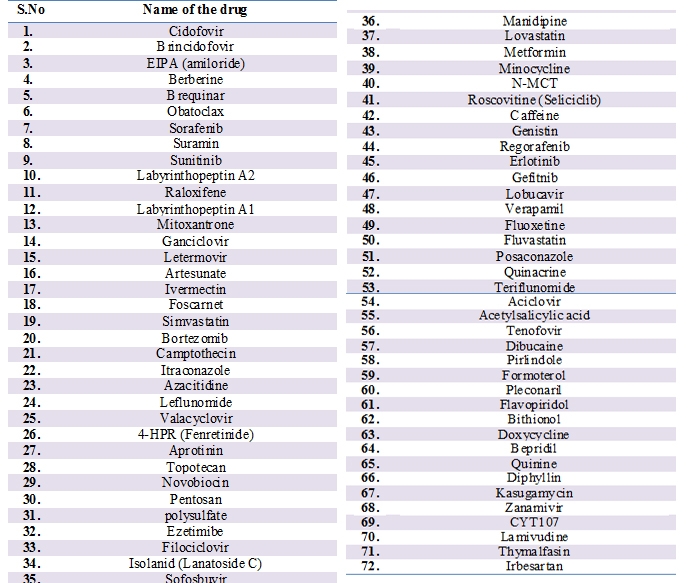
In 2014 Martin Koenighofer et.al, carried out two randomized double blinded controlled trials with 254 virus-positive patients and reported that Carrageenans which are sulfated polysaccharides found in some species of red seaweed based nasal spray showed significant efficacy against human corona virus [39]. In 2014 Adeyemi O. Adedej et.al demonstrated that compound SSYA10-001 blocks SARS-CoV replication by inhibiting its helicase enzyme. They have also evaluated the efficacy of SSYA10-001to inhibit replication of mouse hepatitis virus (MHV) and MERS-CoV and found as potent against them as well [40]. In 2014 Julie Dyall et.al, repurposed clinically developed drugs against MERS-CoV and SARS-CoV. Out of the 290 screened compounds, 27 compounds (Table 1) were found to be active against these two coronaviruses. These compounds belong to different classes of pharmaceuticals, including antipsychotics and anticancer [41].
Table 1: Drugs identified by Canrong Julie Dyall et.al, against MERS-CoV and SARS-CoV:

In 2015 Jianzhong Cao et.al, screened the National Institutes of Health (NIH) Clinical Collection against luciferase reporter-expressing recombinant murine coronavirus in cell culture. A total of 84 compounds were found to have a significant anti-coronavirus effect, with 51 compounds blocking virus entry while 19 others inhibited viral replication, out of the 727 screened. Further validating studies revealed 3 inhibitors (homoharringtonine, nitazoxanide and hexachlorophene) with robust anticoronavirus activities with IC50 ranging from 11 nM to 1.2 μM [42]. In 2015, Hannah L. Peters et.al, designed a series of nucleoside analogues based on the acyclic sugar scaffold of acyclovir and evaluated for their anti-viral potential.
Out of the designed compounds, compound-2 showed significant selective antiviral activity towards HCoV-NL63 and MERS-CoV, but not SARS-CoV interestingly. This study demonstrated that doubly flexible nucleoside analogues can be taken as a new class of drug candidates with potential for selective coronaviruses inhibition [43]. In 2016 Mohammad Reza Dayer et.al, computationally repurposed HIV-1 protease drugs tipranavir, saquinavir, ritonavir, nelfinavir, lopinavir, indinavir, darunavir, atazanavir, and amprenavir onto M protease of coronavirus and reported Lopinavir as potent amongst against Coronavirus Infection [44]. In 2017 Abdo A. Elfiky et.al, reported compounds IDX-184 and MK-0608 as potent inhibitors of human corona viruses targeting RNA-dependent RNA polymerase based on their computational molecular docking studies [45]. In 2018, Christin Muller et.al, reported that silvestrol acts as a potent inhibitor via inhibition of the expression nonstructural and structural proteins of CoV, thus the formation of viral replication complexes. In-vitro inhibitory potential of Silvestrol has been demonstrated on infected human embryonic lung fibroblast (MRC-5) cells with EC50 values of 1.3 nM and 3 nM for MERS-CoV and HCoV-229E, respectively [46]. In 2019, Ayman M. S. Ahmed et.al, reported that 2-Nitrophenylhydrazonopyrazolone derivative 5 showed significant activity against MERS-CoV with EC50 ¼ 4.6 lM [47].
Patents:
In a US patent Olsen DB et.al in 2004, disclosed a nucleoside analogue 4 – amino 7 - (2–C– methyl - β – D -ribofuranosyl ) – 7 Hpyrrolo [ 2 , 3 – d ] pyrimidine as a combination therapy against coronaviruses along with interferon, 2′-C-methylcytidine or inosine monophosphate dehydrogenase inhibitors (levovirin, viramidine, ribavirin) [48]. Fleming RA et.al, in a US patent in 2005 disclosed a phospholipid compound with inhibitory activity against SARS-CoV in Vero cells, as determined by neutral red assay with EC50, CC50 and SI estimated as 3 μg/ml, 40 μg/ml and 13, respectively [49]. Freire E et.al, in a US patent in 2005 disclosed FL-166 a 3CLpro inhibitor derived from biphenyl boronic acid with inhibition constant (Kiapp) of 22 +/- 10 nM [50]. Wu SY et.al, in a US patent in 2006 disclosed one of their designed compounds named Compound (1) had low IC50 inhibitory activity against SARS-CoV 3CLpro [51]. Fujii N et.al, in a US patent in 2006 disclosed that Nelfinavir - HIV-1 protease inhibitor inhibits SARS-CoV 3CLpro with different efficacies by different assays [52]. Pang YP et.al, in a US patent in 2007 disclosed a structurally Tamiflu mimicking molecule CS11 inhibits SARS-CoV 3CLpro with an EC50 of 23 μM (CC50 = 76 μM; SI = 3.3) [53]. Gage PW et.al, in a US patent in 2007 disclosed that Cinnamoylguanidine a “viroporin” inhibitor as a potent coronavirus inhibitor targeting its E protein hindering replication of SARS-CoV, MHV, HCoV-229E and HCoV-OC43 at low μM concentrations [54].
Present:
Drug repurposing approaches
In the present unprecedented situation of deaths on daily basis due to COVID-19, researchers do not have years of time in finding a treatment. In this scenario, repurposing existing drugs with proven safety and toxicology profiles leverage huge advantage in terms of buying time in the process of identifying a potential treatment. Among several advantages of drug repurposing, high probability of success rate, readily available information regarding their synthesis steps, manufacturing processes to foresight regarding different phases of clinical testing are few of the important ones. Moreover in situations like epidemics, repurposing existing medications is the best cut short path to treating those infected by the virus in a novel way. DrugVirus.info database provides manually reviewed information on the discovery and development of broad-spectrum antiviral agents (BSAAs), summarizing activities and developmental statuses of 118 compounds which are safe for humans while targeting 83 human viruses [55]. When this database was queried against HCoV-229E, HCoV-NL63, HCoV-OC43, MERS-CoV, SARS-CoV and novel SARS-CoV-2 a total of fifty one compounds namely were found against COVID-19 infection as possible prophylaxis and treatment candidates. While most of these compounds demonstrated cell culture level antiviral activity, Nitazoxanide, Remdesivir, Emetine and Memantine has been studied upto animal model level. Favipiravir is in Phase II clinical trial. Remdesivir, Hydroxychloroquine and Ritonavir are in advanced stage of Phase III clinical trials against novel SARS-CoV-2 (Figure 4). On March 18, 2020, Shin Watanabe, Michelle Chan and Wataru Suzuki, Nikkei staff writers reported that a recent clinical trial conducted on 200 patients at hospitals in Wuhan and Shenzhen with an influenza medicine favipiravir, is effective against the new coronavirus. Figure 4: Heatmap depicting studies of antiviral compounds against different types of coronaviruses identified so far. Heat map generated via using DrugVirus.info database.
Figure 4: Heatmap depicting studies of antiviral compounds against different types of coronaviruses identified so far. Heat map generated via using DrugVirus.info database.
Patients who were administered with favipiravir tested negative with no side effects, within four days average compared to 11 days in control group [56]. In an another study, researchers who determined the 3CLpro structure repurposed the approved drugs and identified saquinavir, indinavir, lopinavir, ritonavir, carfilzomib along with a schizophrenia medication and two respiratory syncytial virus drugs as potential 3CLpro inhibitors [57].
Malaria drugs for coronavirus treatment
Recent report demonstrated that Chloroquine and hydroxychloroquine as an effective antiviral therapeutic treatment against COVID-19 with faster recovery time. When Azithromycin added to hydroxychloroquine, significant virus elimination was observed. On the other hand, United States Centers for Disease Control and Prevention (US CDC) research shows that chloroquine is a potential prophylactic (preventative) measure against coronavirus. Since, Chloroquine is an inexpensive, globally available drug without any serious side effects against malaria, autoimmune and various other conditions, it’s being considered as one of the hope as immediate treatment or atleast as preventive measure. However, it is still under cautious view; as treating COVID-19 with chloroquine might have fatal side effects in long term. Presently, it is undergoing further validating studies globally. On the other hand, it shall be noted that administration of chloroquine or hydroxychloroquine does have the potential to lead mutation(s) in the virus, which can be either beneficial or harmful to humans [58].
HIV drugs for coronavirus treatment
Although none of the HIV drugs are approved as treatment for coronavirus, based on reports of recovered patients [59-61] many doctors in China, Thailand, Japan and India are administering anti-HIV drugs lopinavir and ritonavir alone and sometimes in combination with other drugs including anti-malarial chloroquine cautiously on case by case basis to treat coronavirus infected patients.
Official guidelines of National Health Commission
According to the guidelines of National Health Commission (NHC) of the People’s Republic of China, antivirals including Interferon Alpha (IFN-α), lopinavir / ritonavir, and ribavirin are recommended for tentative treatment of COVID-19 with recent addition of Chloroquine phosphate and arbidol after their positive preliminary clinical studies (figure 5). The duration of treatment shall not be more than 10 days [62]. Administration of above recommendations for adults is as follows: Vapor inhalation at a dose of 5 million U (and 2 mL of sterile water for injection) 2 times per day is the recommended method of IFN-α administration. lopinavir / ritonavir at a dose of 400 mg/100 mg 2 times per day. Intravenous infusion at a dose of 500 mg Ribavirin in combination with lopinavir / ritonavir or IFN-α 2-3 times per day. Oral administration of Chloroquine phosphate 2 times per day at a dose of 500 mg (300 mg for chloroquine). Oral administration of Umifenovir (Arbidol) 3 times per day at a dose of 200 mg. The patients with high fever exceeding 38.5 °C body temperature can be cautiously administered with common drugs such as ibuprofen 5–10 mg/kg; Paracetamol, also known as acetaminophen 10–15 mg/kg orally [63].

Figure 5: 2D structures of lopinavir, ritonavir, ribavirin, Chloroquine and Umifenovir (Arbidol) drugs.
Future:
Computationally screened compounds worth considering for further studies
As summarized by Falgun Shah et.al, [38] computational tools such as molecular modeling, virtual screening, docking and MD simulations based approaches are proving to be valuable in designing and discovery of target specific compounds with rapid speed, including author’s experience [64-80]. With technological advancements such as increase in computing power and increasing accuracy levels of artificial intelligence and machine learning techniques [81], it is now possible to screen millions of compounds on a specific drug target in a matter of days.
In 2020 Andre Fischer et.al, computationally screened a compound library of over 687 million compounds for binding at the recently solved crystal structure of the main protease of SARS-CoV-2 and report a list of 11 drug-like compounds (CP-1 to CP-11) with improved binding free energy [82]. Canrong Wu et.al, in 2020 employed computational methods and identified few potential compounds against PLpro (Supplementary Table 1), 3CLpro (Supplementary Table 2) and RdRp (Supplementary Table 3) drug targets from ZINC drug database and Natural products database respectively [83].
Yan Li et.al, in 2020 have performed In-silico high-throughput screening based on the 8,000 clinical drug libraries and identified 4 small molecular drugs with high binding capacity namely Prulifloxacin, Bictegravir, Nelfinavir and Tegobuvi against SARS-CoV main protease. Out of these four one used to treat hepatitis C and two aimed at HIV [84]. Xin Liu et.al, in 2020 computationally screened commercially available drugs database from drugbank and have identified 10 small molecular drugs namely, Colistin, Valrubicin, Icatibant, Bepotastine, Epirubicin, Epoprostenol, Vapreotide, Aprepitant, Caspofungin and Perphenazine. These compounds include an anti-nausea medication, an antifungal drug and some cancer-fighting drugs [85]. Richardson, Peter, et.al, in 2020 partnered with BenevolentAI to screen approved drugs using proprietary knowledge graph generated by employing machine learning technique and identified Janus associated kinase (JAK) inhibitor Olumiant (baricitinib), approved for rheumatoid arthritis, on the basis of its inhibition of ACE2-mediated endocytosis. Barcitinib is now being considered for further studies as treatment against COVID-19 [86].
Bowen Tang et.al, in 2020 developed a novel advanced deep Q-learning network with the fragment-based drug design (ADQN-FBDD) based machine learning algorithms to generate 4922 novel lead molecules employing structure-based optimization policy (SBOP) using 3CLpro structure. From the pool of novel compounds thus generated 47 compounds were selected based on their high scoring by AI reward function. Out of the 47 compounds, compound 46 with high covalent docking score was further optimized via implementing chemical biology knowledge and the structure-based optimization policy. All the compounds generated in this study are available at https://github.com/tbwxmu/2019-nCov to undergo synthesis and clinical studies as potential treatment against COVID-19 [87].
Chloroquine analogs with more nitrogens
Based on the positive results of Chloroquine against coronavirus, a group of researchers from Standford University are considering designing chloroquine analogs with more nitrogens. This property of nitrogens to interact with hydrogens makes it harder and harder for the endosome to become acidified, therefore disrupting viral replication. Greater effect of Chloroquine is expected by adding more nitrogens either by increasing the chain length by adding extra carbons and nitrogens around it or by making extra branches of ionizable nitrogens. By doing so, however, alterations in bioavailability and its target specificity is expected [88].
Under clinical trials:
Most of the drugs which are currently under clinical trials are targeted towards coronavirus key components crucial for its lifecycle, starting from viral entry, to its structural and nonstructural proteins responsible for replication, RNA synthesis and release from the cell.
Anti-HIV drugs as a treatment for COVID-19
Abbvie Inc. headquartered at North Chicago, Illinois, United States and Cipla Limited headquartered at Mumbai, India announced that they are studying HIV protease inhibitor, lopinavir along with ritonavir for the treatment of MERS and SARS coronaviruses. Lopinavir is already an approved treatment against HIV infection under trade name Kaletra and Lopimune respectively. During 2003 SARS outbreak, Lopinavir / ritonavir in combination with ribavirin in an open clinical trial showed milder disease course and reduced fatality rate in patients. Keeping in view of these positive results, several clinical trials of lopinavir are underway, either alone or in combination with other drugs such as umifenovir, oseltamivir and baloxavir marboxil. Darunavir, a protease inhibitor approved for HIV-1 treatment along with a boost agent marketed by Janssen Pharmaceutical Companies, a subsidiary of Johnson & Johnson, has donated this drug for further research as a treatment for COVID-19. A group of researchers in China have registered a clinical trial after the anecdotal reports suggests its potential antiviral activity against COVID-19. Chinese drug maker Ascletis Pharma started testing a cocktail of danoprevir and ritonavir, one approved for HIV and another approved for Hepatitis C against COVID-19. Phase-I studies are ongoing with 11 patients enrolled with coronavirus-caused pneumonia for the study. Zhengzhou Granlen PharmaTech registered a clinical trial with Azvudine an experimental reverse transcriptase inhibitor drug against HIV-1/AIDS and testing in combination with marboxil / favipiravir and lopinavir / ritonavir as a potential COVID-19 treatment. When it comes to developing HIV drugs against COVID-19, it is preferable to target virus-specific proteins such as the RNA-dependent RNA polymerase keeping in view of the fact that coronaviruses does not contain or use reverse transcriptase like HIV noted painter [89].
Rheumatoid arthritis drugs against COVID-19
Renmin Hospital of Wuhan University, China had registered a multicenter, randomized, double-blind, controlled phase 3 clinical trial on 200 patients with pneumonia caused by novel coronavirus. randomised to Leflunomide, or placebo. Leflunomide is an EU approved drug for rheumatoid arthritis and psoriasis arthritis [90].
Apart from antiviral drugs, several other registered clinical trials against COVID-19 using a stem cell therapy, immune modulating drugs like glucocorticoids, immunoglobulins, monoclonal antibodies such as sarilumab, tocilizumab and other immune modulating drugs has been summarized in document [91-92].
OYA1 by OyaGen
OyaGen Inc, headquartered at Rochester, New York, United States, recently announced that their compound OYA1 has shown strong antiviral efficacy against live novel coronavirus in cell culture infectivity studies. This compound was earlier approved as investigational new drug against cancer and is presently undertaking further studies regarding its efficacy and safety [93].
Galidesivir, a potential antiviral for coronavirus treatment
Galidesivir (BCX4430) a nucleoside RNA polymerase inhibitor was reported to be shown active against coronavirus via disrupting the viral replication. This compound was previously reported with several benefits against deadly viruses such as Zika, Marburg, Ebola, and Yellow fever. Currently this compound is in advanced stages of in-vitro and in-vivo studies as a broad spectrum antibiotic against coronaviruses, paramyxoviruses, flaviviruses filoviruses, togaviruses, arenaviruses and bunyaviruses [94].
Remdesivir (GS-5734), which was originally developed as an Ebola drug by Gilead Sciences is being studied in five clinical trials with about 1000 patients diagnosed with COVID-19 around the world. Out of this five, two phase-III randomized, placebo-controlled, double-blind clinical trials in Asian countries. According to pilot studies conducted on patients with coronavirus in US, clinical conditions are improved. Further tests regarding safety and efficacy are underway at Nebraska Medical Center [95].
Other compounds
According to a recent study, cellular entry of the SARS-CoV-2 virus has been successfully blocked by TMPRSS2 inhibitor camostat mesylate [96]. Losartan - an AT1 antagonist under the brand name 'Cozaar,' is a common anti-hypertensive agent which is currently prescribed for high blood pressure patients, is being considered against coronavirus based on its biochemical activity such as: converting angiotensinogen to AngI, AngI to AngII (by ACE) [97].
Linlin Zhang et.al in 2020 designed a α-ketoamide inhibitor derivative (compound 13b) based on the crystal structure of SARS-CoV-2 main protease and suggested direct administration of the compound to the lungs would be possible without any adverse effects. In their study, to enhance the half life of the compound in plasma a amide bond was incorporated into pyriodone ring and demonstrated that pyridone-containing inhibitors as a useful framework for development of anti-coronaviral drugs [98].
According to DrugVirus.info database following are the BSAAs which requires further studies to evaluate their potential to act against novel coronavirus (table 2). Since all of these are already approved safe to human drugs, taking these as starting point for design and discovery of SARS-CoV-2 specific drugs would be a worth considering approach.
Table 2: List of Broad-Spectrum Antiviral Agents (BSAAs) which are worth considering for further studies against novel SARS-CoV-2:
Based on the positive results shared by researchers from across globe, WHO have recently launched a global mega trial with most promising Remdesivir, Chloroquine and hydroxychloroquine, Ritonavir / lopinavir and Ritonavir / lopinavir + interferon beta combination to speed up the treatment availability for this pandemic [99].
Conclusion
While the vast information availability about the small molecules with various levels of potential to act as inhibitors against SARS-CoV-2 posses challenges of its own. Leveraging the advanced technologies like artificial intelligence, machine learning, and high throughput screenings does have the potential to speed up the discovery of treatment to a great extent. In pandemic situations like present, it’s a must to form a mastermind with collaborations across globe to access different approaches to solve the problem instead of trying to solve independently. With the ongoing efforts to prevent the spread of 2019-nCoV worldwide, researchers across globe are hopeful that the outbreak may subside in a few months, as with SARS and MERS. Nevertheless, the outbreak has re-emphasized the importance of developing broad-spectrum antiviral agents to combat present and future coronaviruses.
Acknowledgment: Author would like to thank Dr.Mallesh Kurakula, Dr.Iti Kapoor, Dr.Manju Sharan, Dr.Suvarthi Das for generously donating their valuable time in providing peer-review for this manuscript and Md.Rehana Begum for her assistance in typesetting the article.
References
- Sohrabi, Catrin, et al. "World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19)." International Journal of Surgery(2020) doi: 1016/j.ijsu.2020.02.034
- Ruan, YiJun, et al. "Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection." The Lancet9371 (2003): 1779-1785.
- WHO MERS-CoV Research Group. "State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans." PLoS currents5 (2013). doi: 10.1371/currents. outbreaks.0bf719e352e7478f8ad85fa30127ddb8
- Tong, Suxiang, et al. "Detection of novel SARS-like and other coronaviruses in bats from Kenya." Emerging infectious diseases3 (2009): 482.
- Fehr, Anthony R., and Stanley Perlman. "Coronaviruses: an overview of their replication and pathogenesis." Coronaviruses. Humana Press, New York, NY, (2015). 1-23.
- Su, Shuo, et al. "Epidemiology, genetic recombination, and pathogenesis of coronaviruses." Trends in microbiology6 (2016): 490-502.
- Eckerle, Isabella, et al. "Replicative capacity of MERS coronavirus in livestock cell lines." Emerging inf. disease 2 (2014): 276.
- Ren, Li-Li, et al. "Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study." Chinese medical journal(2020) doi: 10.1097/CM9.0000000000000722.
- Yan, Renhong, et al. "Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2." Science(2020) doi: 1126/science.abb2762.
- Ratia, Kiira, et al. "Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme." Proceedings of the National Academy of Sciences15 (2006): 5717-5722.
- Anand, Kanchan, et al. "Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs." Science5626 (2003): 1763-1767.
- Imbert, Isabelle, et al. "A second, non‐canonical RNA‐dependent RNA polymerase in SARS Coronavirus." The EMBO journal20 (2006): 4933-4942.
- Bernini, Andrea, et al. "Tertiary structure prediction of SARS coronavirus helicase." Biochemical and biophysical research communications4 (2006): 1101-1104.
- Decroly, Etienne, et al. "Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex." PLoS pathogens5 (2011). doi: 10.1371/journal.ppat.1002059
- Qiao, Lei, et al. "Discovery, SAR, and X-ray structure of novel biaryl-based dipeptidyl peptidase IV inhibitors." Bioorganic & medicinal chemistry letters1 (2006): 123-128.
- Li, Fang, et al. "Structure of SARS coronavirus spike receptor-binding domain complexed with receptor." Science5742 (2005): 1864-1868.
- Simmons, Graham, et al. "Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry." Proceedings of the National Academy of Sciences33 (2005): 11876-11881.
- Meyer, Daniela, et al. "Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation." Biochemical Journal2 (2013): 331-343.
- Zhai, Yujia, et al. "Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer." Nature structural & molecular biology11 (2005): 980-986.
- Malet, Hélène, et al. "Expression, purification and crystallization of the SARS-CoV macro domain." Acta Crystallographica Section F: Structural Biology and Crystallization Communications4 (2006): 405-408.
- Imbert, Isabelle, et al. "A second, non‐canonical RNA‐dependent RNA polymerase in SARS Coronavirus." The EMBO journal20 (2006): 4933-4942.
- Chen, Ping, et al. "Biochemical characterization of exoribonuclease encoded by SARS coronavirus." BMB Reports5 (2007): 649-655.
- Ricagno, Stefano, et al. "Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family." Proceedings of the National Academy of Sciences32 (2006): 11892-11897.
- Stertz, Silke, et al. "The intracellular sites of early replication and budding of SARS-coronavirus." Virology2 (2007): 304-315.
- Wu, Di, et al. "The SARS-CoV-2 outbreak: what we know." International Journal of Infectious Diseases(2020) doi: 1016/j.ijid.2020.03.004.
- Savarino, Adrea, et al. "Effects of chloroquine on viral infections: an old drug against today's diseases." The Lancet infectious diseases11 (2003): 722-727.
- Vincent, Martin J., et al. "Chloroquine is a potent inhibitor of SARS coronavirus infection and spread." Virology journal1 (2005): 69.
- Cinatl, J., et al. "Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus." The Lancet9374 (2003): 2045-2046.
- Chen, F., et al. "In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds." Journal of Clinical Virology1 (2004): 69-75.
- Jenwitheesuk, Ekachai, and Ram Samudrala. "Identifying inhibitors of the SARS coronavirus proteinase." Bioorganic & Medicinal Chemistry Letters22 (2003): 3989-3992.
- Wu, Chung-Yi, et al. "Small molecules targeting severe acute respiratory syndrome human coronavirus." Proceedings of the National Academy of Sciences27 (2004): 10012-10017.
- Paragas, Jason, et al. "Interferon alfacon1 is an inhibitor of SARS-corona virus in cell-based models." Antiviral research2-3 (2005): 99-102.
- Chen, Lili, et al. "Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro." Journal of virology11 (2005): 7095-7103.
- Yang, Qingang, et al. "Design and synthesis of cinanserin analogs as severe acute respiratory syndrome coronavirus 3CL protease inhibitors." Chemical and Pharmaceutical Bulletin10 (2008): 1400-1405.
- Chen, Xin, Chi-Yuan Chou, and Gu-Gang Chang. "Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme." Antiviral Chemistry and Chemotherapy4 (2009): 151-156.
- Baltina, L. A., et al. "Prospects for the creation of new antiviral drugs based on glycyrrhizic acid and its derivatives (a review)." Pharmaceutical chemistry journal10 (2009):539-548.
- Tong, Tommy R. "Therapies for coronaviruses. Part 2: Inhibitors of intracellular life cycle." Expert opinion on therapeutic patents4 (2009): 415-431.
- Shah, Falgun, et al. "Computational approaches for the discovery of cysteine protease inhibitors against malaria and SARS." Current computer-aided drug design1 (2010): 1-23.
- Koenighofer, Martin, et al. "Carrageenan nasal spray in virus confirmed common cold: individual patient data analysis of two randomized controlled trials." Multidisciplinary respiratory medicine1 (2014): 57.
- Adedeji, Adeyemi O., et al. "Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses." Antimicrobial agents and chemotherapy8 (2014): 4894-4898.
- Dyall, Julie, et al. "Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection." Antimicrobial agents and chemotherapy8 (2014): 4885-4893.
- Cao, Jianzhong, J. Craig Forrest, and Xuming Zhang. "A screen of the NIH Clinical Collection small molecule library identifies potential anti-coronavirus drugs." Antiviral res114 (2015): 1-10.
- Peters, Hannah L., et al. "Design, synthesis and evaluation of a series of acyclic fleximer nucleoside analogues with anti-coronavirus activity." Bioorganic & medicinal chemistry letters15 (2015): 2923-2926.
- Dayer, Mohammad Reza, Sara Taleb-Gassabi, and Mohammad Saaid Dayer. "Lopinavir; A Potent Drug against Coronavirus Infection: Insight from Molecular Docking Study." Archives of Clinical Infectious Diseases4 (2017). e13823
- Elfiky, Abdo A., Samah M. Mahdy, and Wael M. Elshemey. "Quantitative structure‐activity relationship and molecular docking revealed a potency of anti‐hepatitis C virus drugs against human corona viruses." Journal of medical virology6 (2017): 1040-1047.
- Müller, Christin, et al. "Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona-and picornaviruses." Antiviral research150 (2018): 123-129.
- Ahmed, Ayman MS, et al. "3-Trifluoromethylpyrazolones derived nucleosides: Synthesis and antiviral evaluation." Nucleosides, Nucleotides and Nucleic Acids8 (2019): 590-603.
- Olsen, David, et al. "Inhibiting Coronaviridae viral replication and treating Coronaviridae viral infection with nucleoside compounds." U.S. Patent Application No. 10/832,945.
- Fleming, Ronald, et al. "Phospholipids for the treatment of infection by togaviruses, herpes viruses and coronaviruses." U.S. Patent Application No. 10/783,927.
- Freire, Ernesto, et al. "Inhibitors of coronavirus protease and methods of use thereof." U.S. Patent Application No. 10/979,871.
- Wu, Su-Ying, et al. "SARS CoV main protease inhibitors." U.S. Patent Application No. 11/185,917.
- Fujii N, Yamamoto N, “Anti-coronavirus drug” S. Patent Application No 20060223847; 2006.
- Pang, Yuan-Ping, Andrea Dooley, and Jewn Park. "Antiviral Compositions and Methods." U.S. Patent Application No. 11/557,396.
- Gage, Peter, et al. "Antiviral compounds and methods." U.S. Patent Application No. 10/562,296.
- Andersen, Petter I., et al. "Discovery and development of safe-in-man broad-spectrum antiviral agents." International Journal of Infectious Diseases(2020). doi: 10.1016/j.ijid.2020.02.018.
- Shin Watanabe, Michelle Chan and Wataru Suzuki “China says Japan-developed drug Avigan works against coronavirus” Nikkei. Available at : https://asia.nikkei.com/Business/Pharmaceuticals/China-says-Japan-developed-drug-Avigan-works-against-coronavirus2
- Charlotte Harrison: Coronavirus puts drug repurposing on the fast Track (2020). doi: 10.1038/d41587-020-00003-1
- Gautret, Philippe, et al. "Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial." International Journal of Antimicrobial Agents(2020): 105949.
- Tamar Lapin, HIV drugs are being used as part of coronavirus treatment march 12 2020 NYpost. Avaialbe online at: https://nypost.com/2020/03/12/hiv-drugs-are-being-used-as-part-of-coronavirus-treatment/
- Rajasthan doctors cure coronavirus patient with HIV drugs March 21 2020 Available online at:
http://timesofindia.indiatimes.com/ articleshow/74584859.cms?utm_source=contentofinterest&utm_medium=text&utm_campaign=cppst - Coronavirus pandemic: Foreigner treated with HIV drugs for Covid-19 tests negative March 26 2020. Available online at: https://www.indiatoday.in/india/story/coronavirus-pandemic-foreigner-treated-hiv-drugs-covid-19-tests-negative-1659761-2020-03-26
- Dong, Liying, Shasha Hu, and Jianjun Gao. "Discovering drugs to treat coronavirus disease 2019 (COVID-19)." Drug Discoveries & Therapeutics1 (2020): 58-60.
- Shen, Kunling, et al. "Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: experts’ consensus statement." World Journal of Pediatrics(2020): 1-9.
- Basha, Syed Hussain, and R. Nalini Prasad. "In-Silico screening of Pleconaril and its novel substituted derivatives with Neuraminidase of H1N1 Influenza strain." BMC research notes1 (2012): 105.
- Hussain Basha, S., and K. Naresh Kumar. "Ligand and Structure based virtual screening studies to identify potent inhibitors against herpes virus targeting gB-gH-gL complex interface as a novel drug target." Open access sci rep12 (2012): 566.
- Basha, Syed Hussain, Deepthi Talluri, and Nalini Prasad Raminni. "Computational repositioning of ethno medicine elucidated gB-gH-gL complex as novel anti herpes drug target." BMC complementary and alternative medicine1 (2013): 1-11.
- Basha, Syed Hussain, P. Bethapudi, and F. Majji Rambabu. "Anti-angiogenesis property by Quercetin compound targeting VEGFR2 elucidated in a computational approach." European Journal of Biotechnology and Bioscience6 (2014): 30-46.
- Rao, Chennu Maruthi Malya Prasada, et al. "Molecular docking based screening of novel designed chalcone series of compounds for their anti-cancer activity targeting EGFR kinase domain." Bioinformation7 (2015): 322.
- Reddy, S. V. G., et al. "Molecular docking and dynamic simulation studies evidenced plausible immunotherapeutic anticancer property by Withaferin A targeting indoleamine 2, 3-dioxygenase." Journal of Biomolecular Structure and Dynamics12 (2015): 2695-2709.
- Basha, Syed Hussain, Abhishek Thakur, and Firoz A. Samad. "Insights from the predicted structural analysis of carborane substituted withaferin A with Indoleamine-2, 3-dioxygenase as a potent inhibitor." Bioinformation9 (2016): 374.
- Abdul, F. Samad, et al. "A Comprehensive In Silico Analysis on the Structural and Functional Impact of SNPs in the Congenital Heart Defects Associated with NKX2-5 Gene-A Molecular Dynamic Simulation Approach." PloS one5 (2016): e0153999.
- Chauhan, Navneet, Anuradha Gajjar, and Syed Hussain Basha. "Pharmacophore feature-based virtual screening for finding potent GSK-3 inhibitors using molecular docking and dynamics simulations." Bioinformation10 (2016): 391.
- Wakure, B. S., N. M. Bhatia, and Hussain Basha Syed. "Noninvasive cellular internalization of silver molecules by chitosan nanoneedles: a novel nanocarrier." Journal of Biomolecular Structure and Dynamics5 (2016): 971-982.
- Rather, Mohd Ashraf, et al. "Characterization, molecular docking, dynamics simulation and metadynamics of kisspeptin receptor with kisspeptin." International journal of biological macromolecules101 (2017): 241-253.
- Timiri, Ajay Kumar, et al. "In silico development of a novel putative inhibitor of the 3C protease of Coxsackievirus B3 with a benzene sulfonamide skeleton." Journal of Pharmaceutical Chemistry3 (2017): 25-34.
- Murthy, NVS Viswanadha. "M, V. Girija Sastry, Syed Hussain Basha. 3, 5-dinitrophenyl clubbed azoles against latent tuberculosis-a theoretical mechanistic study." Journal of PeerScientist1 (2018): e1000001.
- Khan, Shah Alam, et al. "Synthesis, molecular docking with COX 1& II enzyme, ADMET screening and in vivo anti-inflammatory activity of oxadiazole, thiadiazole and triazole analogs of felbinac." Journal of Saudi Chemical Society4 (2018): 469-484.
- Krishnaveni, M., and Syed Hussain Basha. "Impact of SNP mutations on the structural and functional behaviour of CD44 associated with oral cancer." International Journal of Pharmacy and Biological Sciences, 2(2018) 352-364.
- Rao, CH MM Prasada. "Novel series of 1, 5 Benzothiazepine skeleton based compounds as anti-cancer agents–In silico and MTT assay based study." Journal of PeerScientist2 (2018): e1000008.
- Ali, M. Ajmal, et al. "In Silico Elucidation of the Plausible Inhibitory Potential of Withaferin A of Withania Somnifera Medicinal Herb Against Breast Cancer Targeting Estrogen Receptor." Current Pharmaceutical Biotechnology(2020) doi : 10.2174/1389201021666200129121843.
- Banerjee, Amit Kumar, and Neelima Arora. "Machine learning techniques in biological data classification and clustering: Initiation of a scientific voyage." Journal of PeerScientist1 (2020): e1000011.
- Fischer, André, et al. "Inhibitors for Novel Coronavirus Protease Identified by Virtual Screening of 687 Million Compounds." (2020) preprint. doi : 10.26434/chemrxiv.11923239.v1
- Wu, Canrong, et al. "Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods." Acta Pharmaceutica Sinica B(2020) doi: 10.1016/j.apsb.2020.02.008.
- Li, Yan, et al. "Therapeutic Drugs Targeting 2019-nCoV Main Protease by High-Throughput Screening." bioRxiv(2020). Doi: 11101/2020.01.28.922922
- Liu, Xin, and Xiu-Jie Wang. "Potential inhibitors for 2019-nCoV coronavirus M protease from clinically approved medicines." bioRxiv(2020). Doi: 10.1101/2020.01.29.924100
- Richardson, Peter, et al. "Baricitinib as potential treatment for 2019-nCoV acute respiratory disease." The Lancet10223 (2020): e30-e31.
- Tang, Bowen, et al. "AI-aided design of novel targeted covalent inhibitors against SARS-CoV-2." bioRxiv(2020) doi: 1101/2020.03.03.972133.
- James M. Todaro, Gregory J. Rigano: An Effective Treatment for Coronavirus (COVID-19) March 13, 2020, 1-15
- Painter, Meghan M., et al. "Antiviral protection via RdRP-mediated stable activation of innate immunity." PLoS pathogens12 (2015). doi : 10.1371/journal.ppat.1005311
- Fox, Robert I., et al. "Mechanism of action for leflunomide in rheumatoid arthritis." Clinical Immunology3 (1999): 198-208.
- Overview of planned or ongoing studies of drugs for the treatment of COVID-19 27.03.2020 https://laegemiddelstyrelsen.dk/ da/nyheder/temaer/ny-coronavirus-covid-19/~/media/ 5B83D25935DF43A38FF823E24604AC36.ashx
- Singh, Awadhesh Kumar, et al. "Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: A systematic search and a narrative review with a special reference to India and other developing countries." Diabetes & Metabolic Syndrome: Clinical Research & Reviews(2020).doi 1016/j.dsx.2020.03.011
- OyaGen, Inc. Announces a Compound in Development with Broad Antiviral Activity Against Coronaviruses, including SARS-CoV-2 https://www.prnewswire.com/news-releases/oyagen-inc-announces-a-compound-in-development-with-broad-antiviral-activity-against-coronaviruses-including-sars-cov-2-301021543. html
- Rele, Shyam. "Emerging outbreaks and epidemic threats: The practicality and limitations in the development and manufacturing of treatments for Coronavirus (COVID-19)." Polymorphism4 (2020): 45-52.
- Li, Qun, et al. "Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia." New England Journal of Medicine13(2020). 1199-1207.
- Hoffmann, Markus, et al. "SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor." Cell(2020) doi: 1016/j.cell.2020.02.052
- Zeinalian, Mehrdad, et al. "A potential protective role of Losartan against coronavirus induced lung damage." Infection Control & Hospital Epidemiology: 1-6. doi: 10.1017/ice.2020.80
- Zhang, Linlin, et al. "X-ray Structure of Main Protease of the Novel Coronavirus SARS-CoV-2 Enables Design of α-Ketoamide Inhibitors." bioRxiv(2020). doi: 10.1101/2020.02.17.952879
- Tung, H. Y. "In The Matter Of The Virus That Causes Corona Virus Disease (COVID-19), SARS-COV-2: A Case Predicted By The Theory Of Chaos." ScienceOpen Preprints(2020) doi: 10.14293/S2199-1006.1.sor-.ppmh5lh.v1.